Interactive Platform for Synthetic Flow Cytometry Data Generation for Education
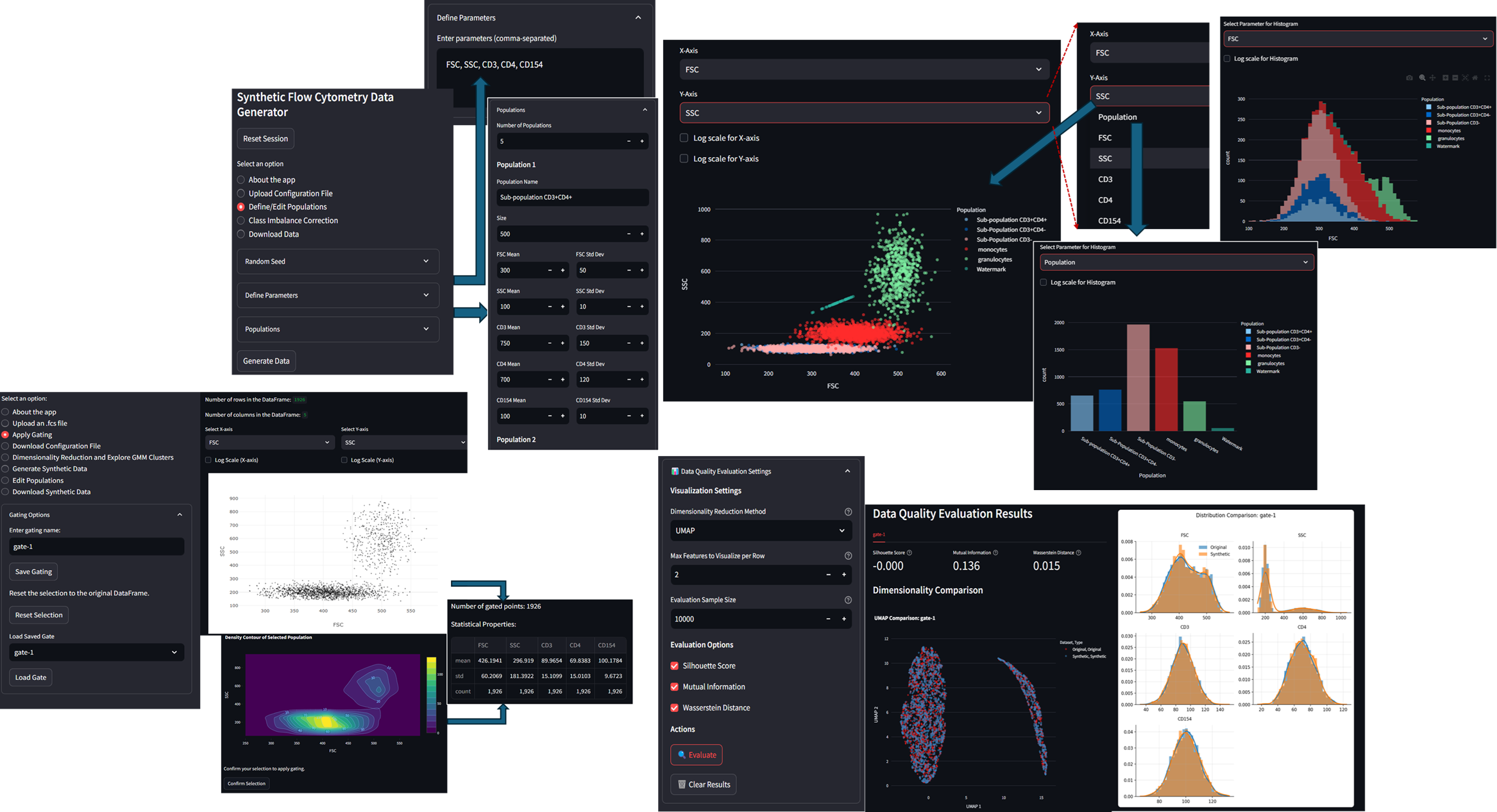
Developed web-based platform to generate and analyze synthetic flow cytometry (FCM)
data for educational use, addressing challenges in FCM education; data accessibility, privacy, and complexity of real FCM datasets by providing customizable, noise-free synthetic
alternatives that simulate real-world biological structures where these can be used for teaching core analytical concepts like gating and clustering.
Two applications built using Streamlit and Python, emphasize user-friendliness, statistical rigor, rapid prototyping, reproducibility and integration with existing FCM workflows.
- App-1: Allows users to generate synthetic FCM data from scratch using Gaussian distributions. Users can define population characteristics, control population compositions, class imbalance, and apply watermarks for traceability.
- App-2: Extracts cell populations from real FCM files using Gaussian Mixture Models (GMM), with dimensionality reduction (UMAP/t-SNE/PCA), enabling dynamic resampling and visualization. It supports dynamic resampling, includes validation metrics (Silhouette Score, Mutual Information, Wasserstein Distance) to compare synthetic data against original datasets.
A brief demonstration for App-1:
A brief demonstration for App-2:
Impact:
Both apps offer real-time data visualization, export to FCS/CSV, and tools for editing cell populations interactively, enhancing FCM education by providing clean, customizable datasets while maintaining statistical rigor through validation metrics.
Designed to support educational use by simplifying key concepts like gating and clustering, while also having the potential to extend its usage in FCM research and clinical applications for benchmarking computational tools .
Key features include:
- User-friendly UI for defining or extracting cell populations
- Statistical validation of synthetic data
- Support for configuration sharing and reproducibility
- Simulated experimental variability
Future improvements may include non-Gaussian models and expanded metadata support for higher biological fidelity.
Software/packages used:
Python, Streamlit, Scikit-learn (GMM, t-SNE, UMAP, PCA, mutual_info_score, silhouette_score), NumPy, Pandas, Plotly, Matplotlib, SciPy (wasserstein_distance)