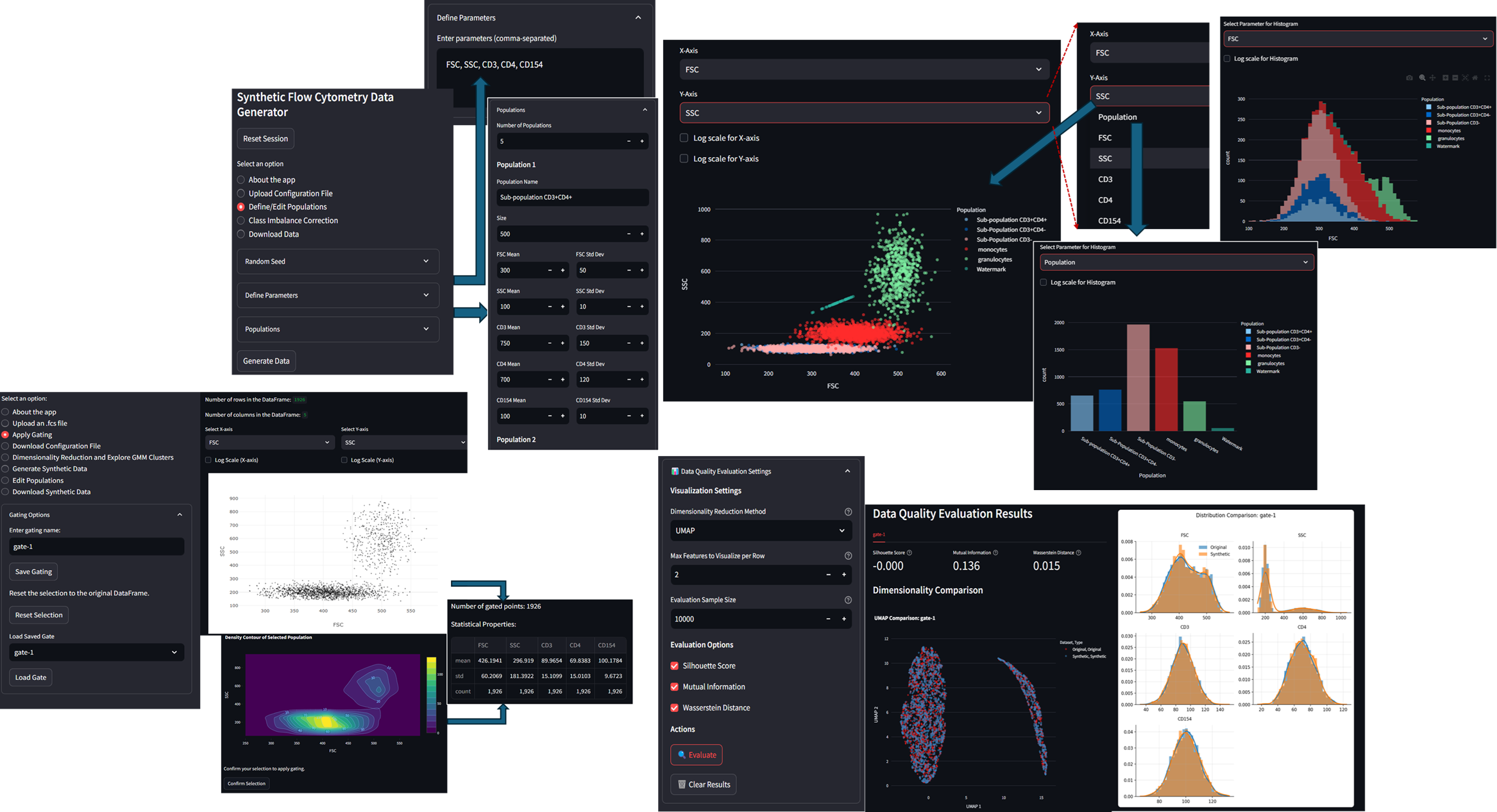
Developed web-based platforms to generate and analyze synthetic flow cytometry (FCM)
data for educational use ..
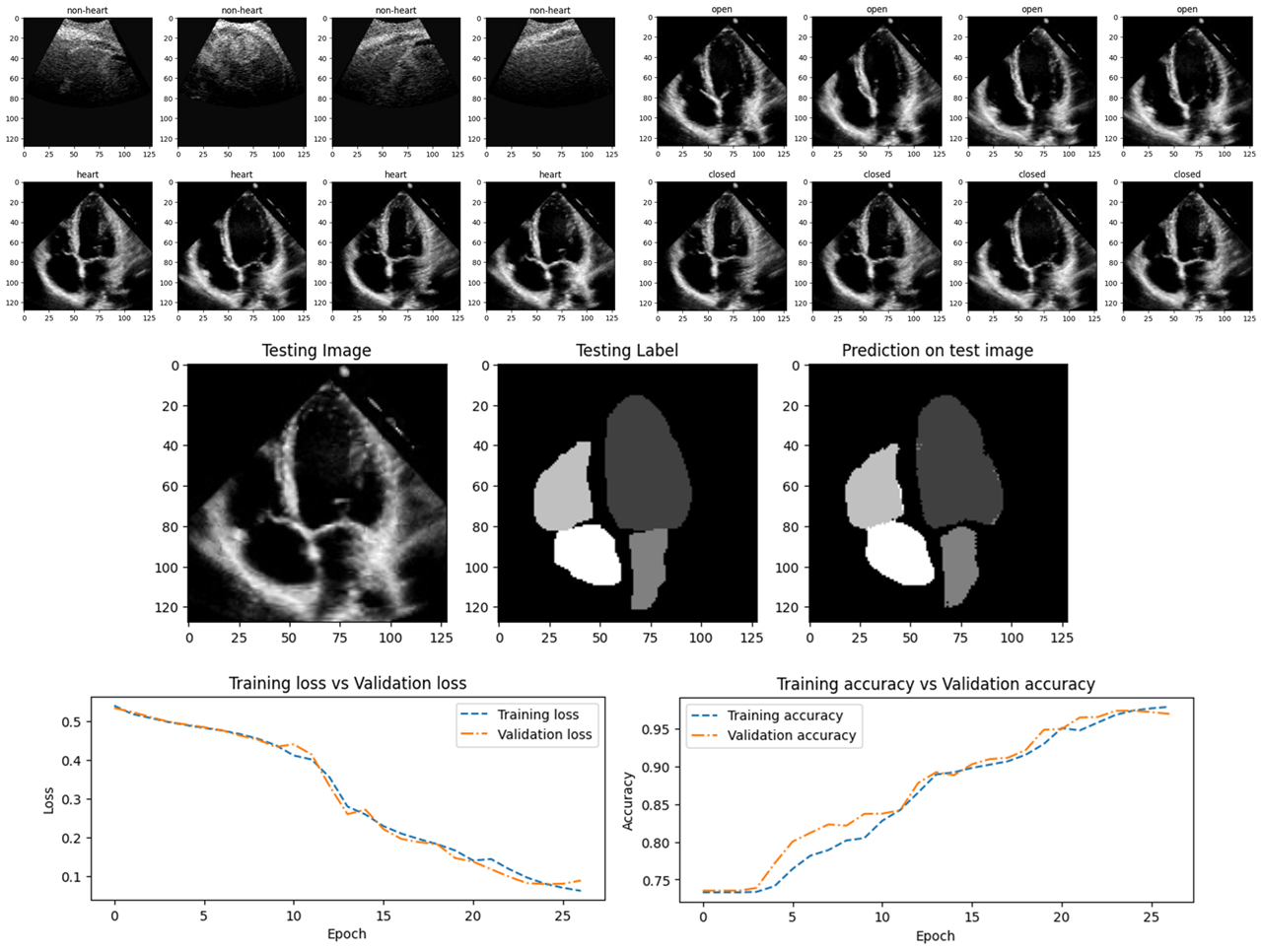
Developed machine learning and deep learning models to classify and interpret ultrasound images for clinical decision support.
The coursework was completed in two stages:
Heart vs. Abdominal Ultrasound Classification:
- Classify heart vs abdominal ultrasound images using SVM and CNN models, achieving 100% accuracy on test data using both model architectures.
- Implemented 5-fold cross-validation and analyzed performance via precision, recall, F1-score, and ROC-AUC (all 1.0).
Heart Valve State Detection (Open vs. Closed Mitral Valve):
- Designed a custom CNN architecture (3 Convolutional layers, dropout, and dense layers), achieving 97.7% test accuracy (AUC: 0.97).
- Addressed class imbalance via stratified sampling and early stopping, with 94.9 ± 2.8 % mean accuracy in 5-fold CV.
Performance Optimization
- Improved CNN generalization using data augmentation (rotation, zoom), boosting test AUC to 1.0 (perfect separation).
- Applied transfer learning with ResNet152V2 (fine-tuned last 20 layers), increasing AUC from 0.84 (frozen layers) to 0.91 (fine-tuned).
Clinical insight via Segmentation
- Implemented a U-Net model to segment heart chambers from clinician-annotated masks, achieving 92.8% validation accuracy and quantifying overlap via Intersection over Union (IoU).
- Visualized predictions against ground truth for interpretability.
- Segmentation outputs enhanced interpretability in valve classification tasks.
Key contributions:
Demonstrated end-to-end ML pipeline: data loading, preprocessing, model design, evaluation, and deployment-ready saving (pickle).
Leveraged augmentation and transfer learning to enhance robustness, with clear metrics-driven validation.
Provided clinical insight via segmentation, enabling precise anatomical context for heart valve state classification.
Software/packages:
Python, TensorFlow, Keras, Scikit-learn, OpenCV, NumPy, Pandas, Matplotlib, Seaborn, scikit-image, PIL (Pillow), imutils
This was conducted as part of an academic module at RGU, where the goal was to apply machine learning techniques to medical imaging data classification and segmentation tasks.
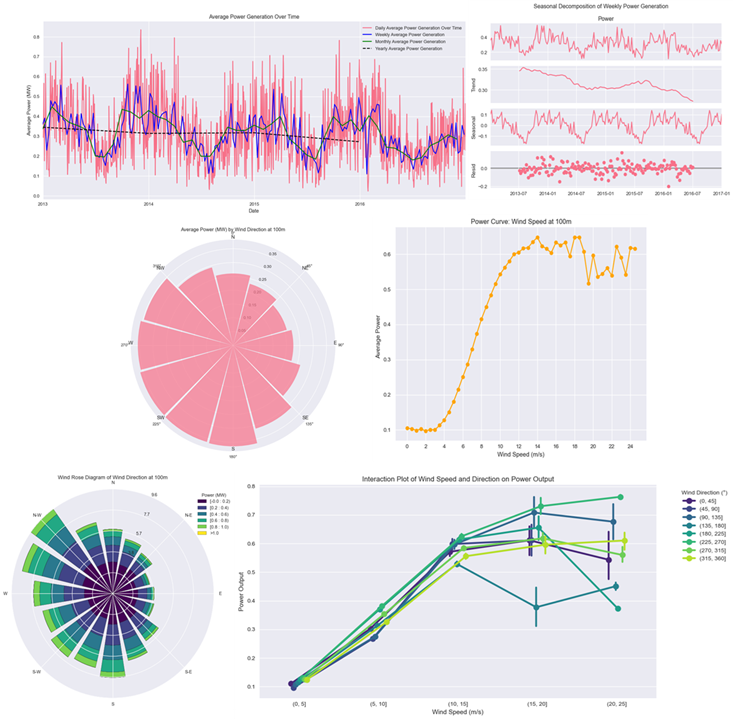
This project presents an in-depth Exploratory Data Analysis (EDA) of wind turbine power generation data.
Part 1
- This part aims to uncover the key patterns, trends, and relationships between features such as wind speed, direction, and generated power.
- The analysis includes visualizations of time series data, seasonal trends, and correlations between features.
- It also explores the impact of weather conditions on power generation, identifying optimal wind conditions for maximum output.
- Includes visual analytics, domain insights, and groundwork for predictive modelling.
Part 2
- aims at training a machine learning model to predict power generation based on available features.
Software/packages used:
Python, Pandas, NumPy, Matplotlib, Seaborn, Scikit-learn



